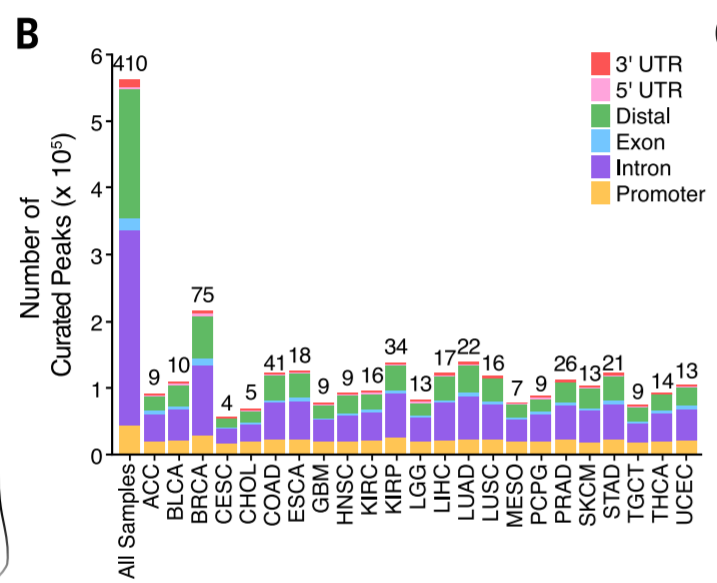
- Peak distribution
Peak calling + annotate distribution
2. Signal track
Calculate coverage + normalization
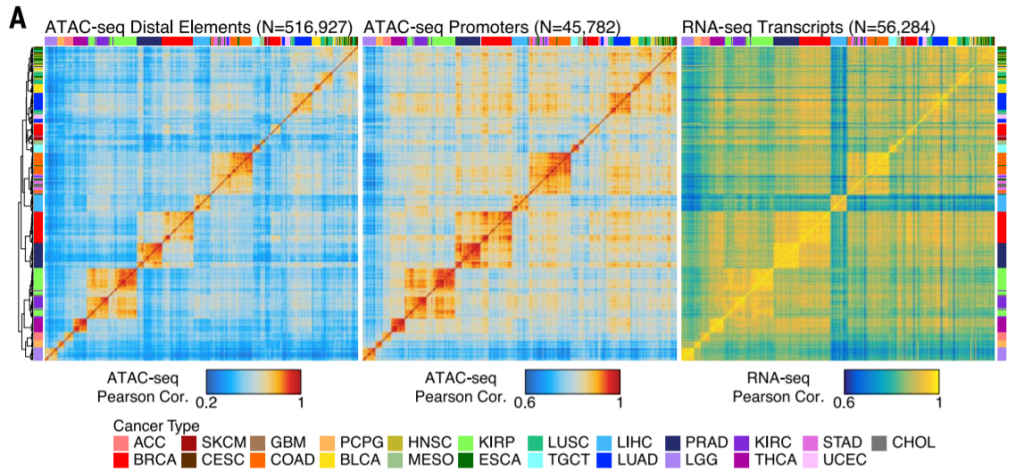
3. Pearson correlation heatmap
Large sample size + split component (distal/promoter) + calculate Pearson correlation
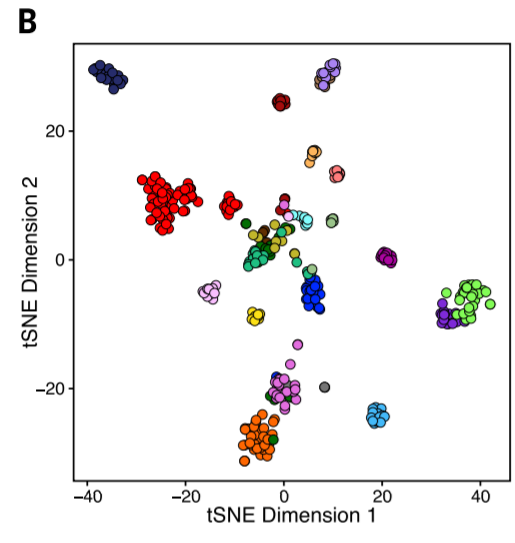
4. Unsupervised tSNE clustering
Large sample size + tSNE
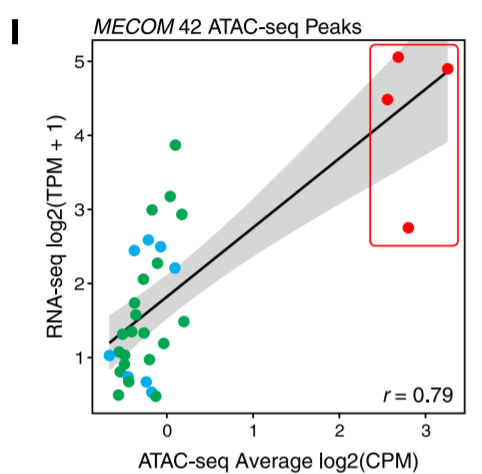
5. Correlation plot of chromatin accessibility and gene expression
Large sample size + RNA-seq + linear regression
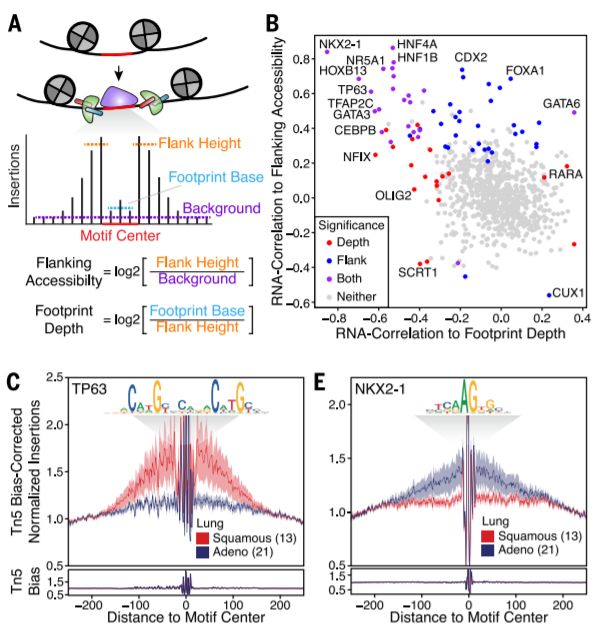
6. TF footprinting analysis
Require sequencing depth + long calculation time
(B): + RNA-seq
❗ ❗ not compitable with motif enrichment
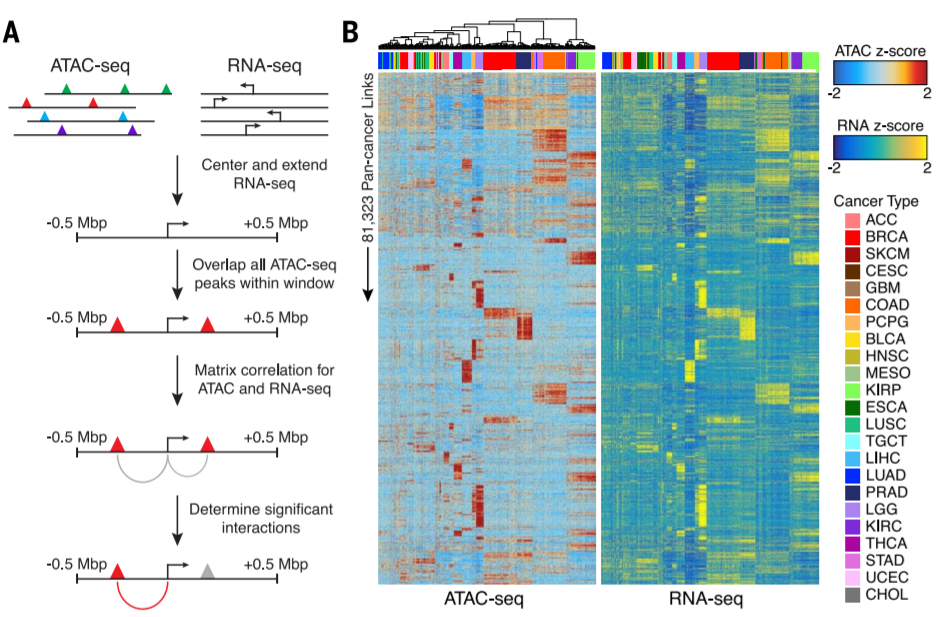
7. ATAC-seq peaks to genes link prediction
Large sample size + ATAC-eQTL
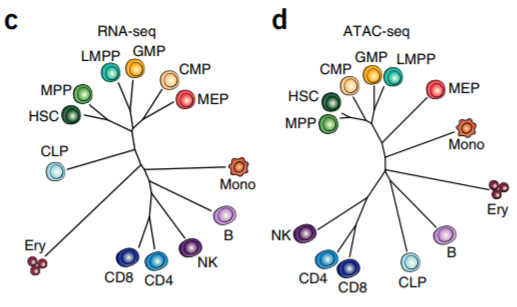
8. Phylogenetic dendrogram
Large sample size + clustering
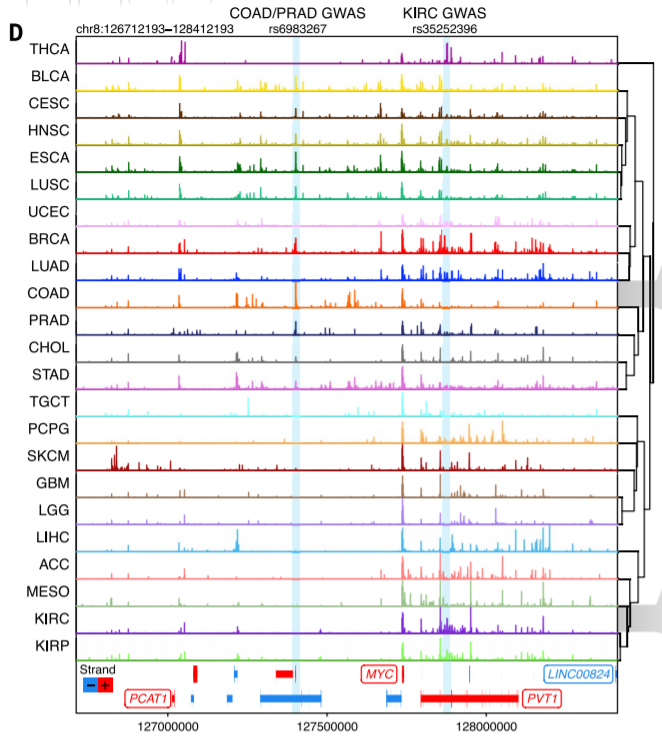
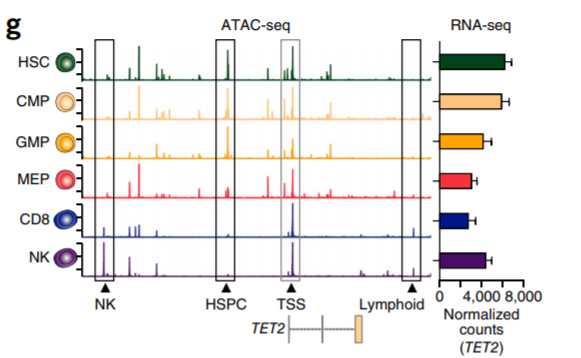
9. Signal track and gene expression
Compute coverage + normalization + RNA-seq
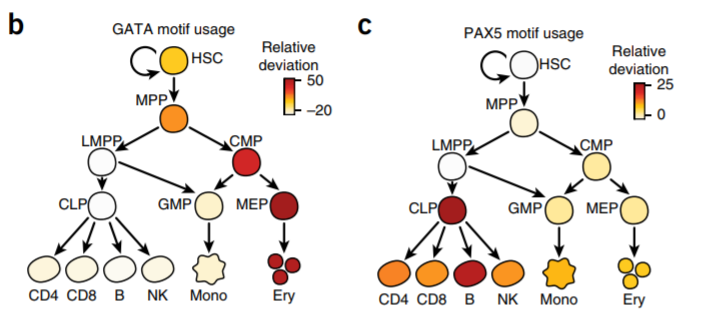
10. TF usage across crowds
Large sample size/scATAC-seq + motif enrichment (chromVAR)
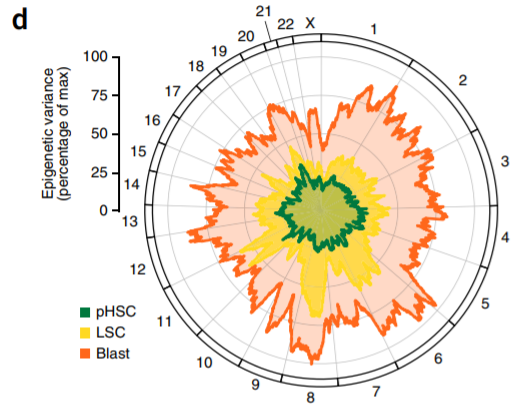
11. Mean variance in ATAC-seq signal across the linear genome
Large sample size + calculate variance
12. Cell type components deconvolution
Require established signature + suport vector regression (CIBERSORT)
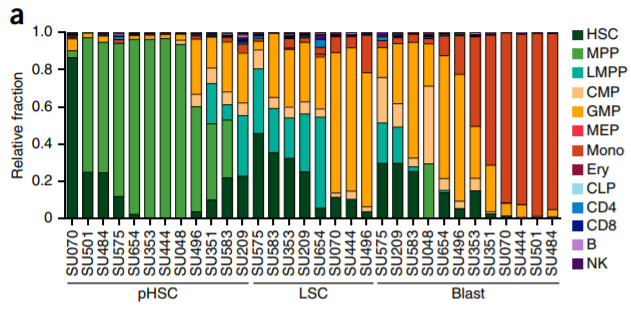
13. Peak accessibility changes
Chronological samples (#bars = sample size - 1)
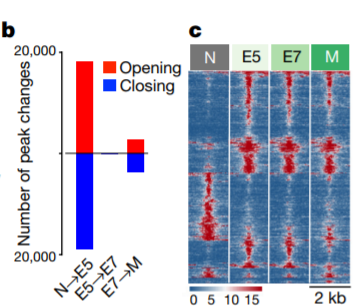
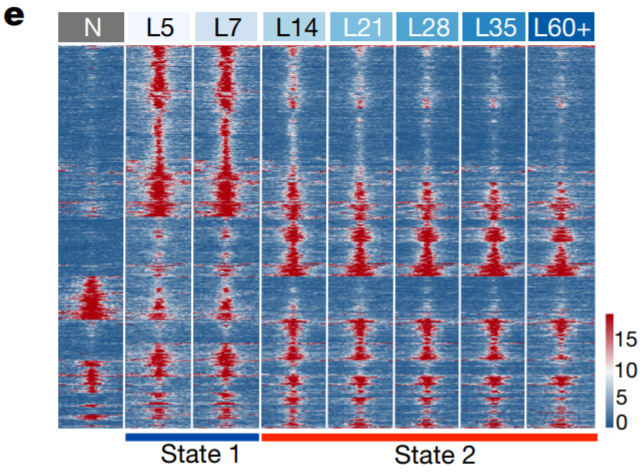
14. Chromatin accessibility heatmap
Compute coverage
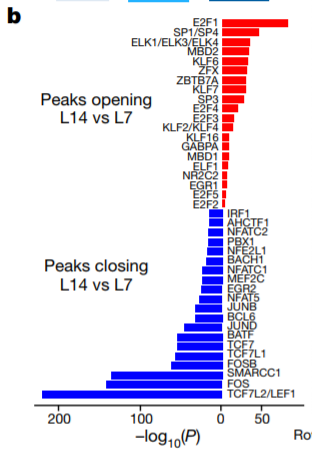
15. TF motif enrichment between 2 samples
Differential peak calling + motif enrichment
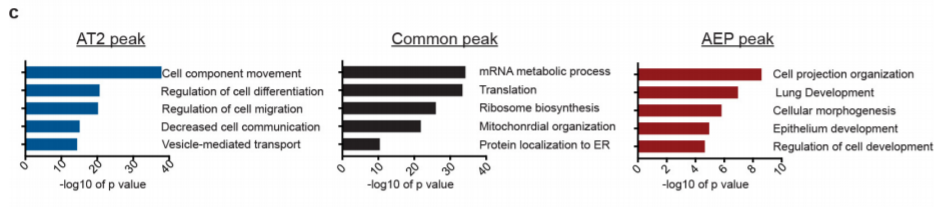
16. GO enrichment of peak nearby genes
Peak calling + GO enrichment
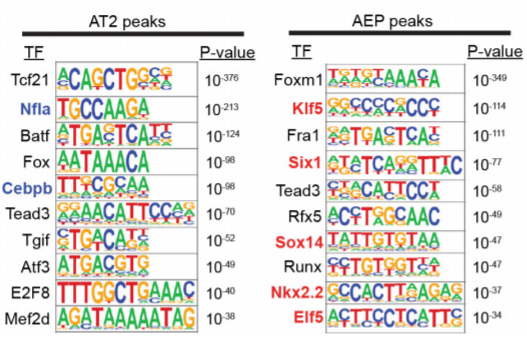
17. TF motif enrichment
peak calling + motif enrichment
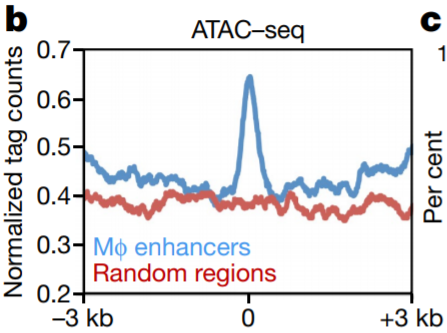
18. Singal density plot
Compute coverage + calculate average density
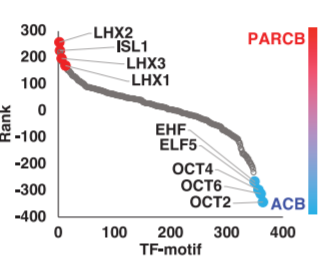
19. TF motif enrichment downfall plot
Peak calling + motif enrichment
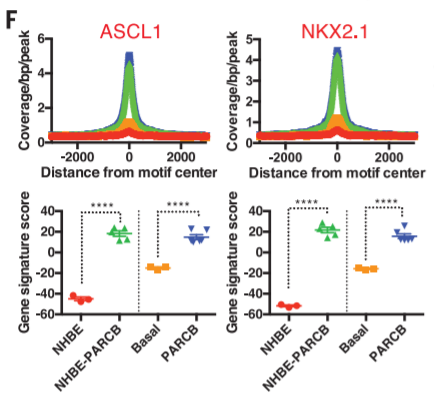
20. TF 'footprint' + gene signature score
Peak calling + footprint + RNA-seq
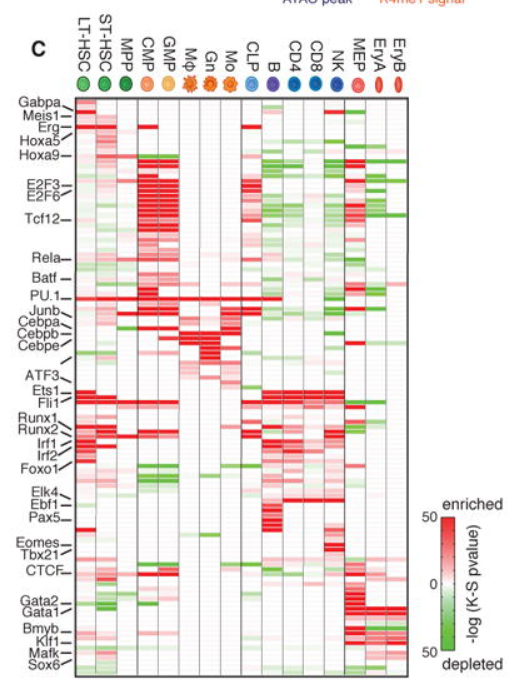
21. TF heatmap
Peak calling + motif enrichment + map P value